Tutorial 8: CV preset
Official LightAutoML github repository is here
In this tutorial we will look how to apply LightAutoML to computer vision tasks.
Basically, the corresponding modules are designed to solve problems where the image is more of an auxiliary value (complement the rest of the data from the table) than for solving full-fledged CV problems. In LightAutoML working with images goes essentially through tabular data, that is, not the images themselves are used, but the paths for them. They should be written in a separate column, which needs to specify the corresponding 'path' role. The target variable and optionally other
features are also specified in the table. To make predictions, numerical features are extracted from images, such as color histograms (RGB or HSV) and image embeddings based on EfficientNet (with the option to select a version and use AdvProp weights), and then standard machine learning models available in LightAutoML (as in conventional tabular presets) can be applied to them. By default, linear regression with L2 regularization and CatBoost are used.
Linear regression is trained on image embeddings, CatBoost is trained on histogram features, and weighted blending is finally applied to their predictions.
As an example, let’s consider the Paddy Doctor competition - the task of multi-class classification, determining the type of paddy leaf disease based on photographs and other numerical features. Data is a set of images and a table, each row of which corresponds to a specific image with a specification of the path to it.
Importing libraries and preparing data
We will use the data from Kaggle. You can download the dataset from this link and import it in any convenient way. For example, we download the data using kaggle API and install some corresponding requirements. You can run next cell for loading data and installing packages in this way:
[ ]:
##Kaggle functionality for loading data; Note that you have to use your kaggle API token (see the link above):
# !pip install opendatasets
# !pip install -q kaggle
# !pip install --upgrade --force-reinstall --no-deps kaggle
# !mkdir ~/.kaggle
# !ls ~/.kaggle
# !cp kaggle.json ~/.kaggle/
# !chmod 600 ~/.kaggle/kaggle.json
# !kaggle competitions download -c paddy-disease-classification
# #Unpack data:
# !mkdir paddy-disease
# !unzip paddy-disease-classification.zip -d paddy-disease
# #Install LightAutoML, Pandas and torch EfficientNet:
# !pip install -U lightautoml[cv] #[cv] is for installing CV tasks functionality
Then we will import the libraries we use in this kernel: - Standard python libraries for timing, working with OS etc. - Essential python DS libraries like numpy, pandas, scikit-learn and torch (the last we will use in the next cell) - LightAutoML modules: TabularCVAutoML preset for AutoML model creation and Task class to setup what kind of ML problem we solve (binary/multiclass classification or regression)
[1]:
import os
os.environ["CUDA_DEVICE_ORDER"]="PCI_BUS_ID" # see issue #152
os.environ["CUDA_VISIBLE_DEVICES"]="0"
[2]:
# Standard python libraries
import os
import time
# Essential DS libraries
import numpy as np
import pandas as pd
from sklearn.metrics import f1_score, accuracy_score, confusion_matrix
from sklearn.model_selection import train_test_split
import torch
import seaborn as sns
import matplotlib.pyplot as plt
# LightAutoML presets, task and report generation
from lightautoml.automl.presets.image_presets import TabularCVAutoML
from lightautoml.tasks import Task
'nlp' extra dependecy package 'gensim' isn't installed. Look at README.md in repo 'LightAutoML' for installation instructions.
'nlp' extra dependecy package 'nltk' isn't installed. Look at README.md in repo 'LightAutoML' for installation instructions.
'nlp' extra dependecy package 'transformers' isn't installed. Look at README.md in repo 'LightAutoML' for installation instructions.
'nlp' extra dependecy package 'gensim' isn't installed. Look at README.md in repo 'LightAutoML' for installation instructions.
'nlp' extra dependecy package 'nltk' isn't installed. Look at README.md in repo 'LightAutoML' for installation instructions.
'nlp' extra dependecy package 'transformers' isn't installed. Look at README.md in repo 'LightAutoML' for installation instructions.
/home/dvladimirvasilyev/LightAutoML/lightautoml/ml_algo/dl_model.py:41: UserWarning: 'transformers' - package isn't installed
warnings.warn("'transformers' - package isn't installed")
/home/dvladimirvasilyev/LightAutoML/lightautoml/text/nn_model.py:22: UserWarning: 'transformers' - package isn't installed
warnings.warn("'transformers' - package isn't installed")
/home/dvladimirvasilyev/LightAutoML/lightautoml/text/dl_transformers.py:25: UserWarning: 'transformers' - package isn't installed
warnings.warn("'transformers' - package isn't installed")
For better reproducibility fix numpy random seed with max number of threads for Torch (which usually try to use all the threads on server):
[3]:
np.random.seed(42)
torch.set_num_threads(2)
Let’s check the data we have:
[4]:
INPUT_DIR = './paddy-disease/'
[5]:
train_data = pd.read_csv(INPUT_DIR + 'train.csv')
print(train_data.shape)
train_data.head()
(10407, 4)
[5]:
| image_id | label | variety | age | |
|---|---|---|---|---|
| 0 | 100330.jpg | bacterial_leaf_blight | ADT45 | 45 |
| 1 | 100365.jpg | bacterial_leaf_blight | ADT45 | 45 |
| 2 | 100382.jpg | bacterial_leaf_blight | ADT45 | 45 |
| 3 | 100632.jpg | bacterial_leaf_blight | ADT45 | 45 |
| 4 | 101918.jpg | bacterial_leaf_blight | ADT45 | 45 |
[6]:
train_data['label'].value_counts()
[6]:
normal 1764
blast 1738
hispa 1594
dead_heart 1442
tungro 1088
brown_spot 965
downy_mildew 620
bacterial_leaf_blight 479
bacterial_leaf_streak 380
bacterial_panicle_blight 337
Name: label, dtype: int64
[7]:
train_data['variety'].value_counts()
[7]:
ADT45 6992
KarnatakaPonni 988
Ponni 657
AtchayaPonni 461
Zonal 399
AndraPonni 377
Onthanel 351
IR20 114
RR 36
Surya 32
Name: variety, dtype: int64
[8]:
train_data['age'].value_counts()
[8]:
70 3077
60 1660
50 1066
75 866
65 774
55 563
72 552
45 505
67 415
68 253
80 225
57 213
47 112
77 42
73 38
66 36
62 5
82 5
Name: age, dtype: int64
[9]:
submission = pd.read_csv(INPUT_DIR + 'sample_submission.csv')
print(submission.shape)
submission.head()
(3469, 2)
[9]:
| image_id | label | |
|---|---|---|
| 0 | 200001.jpg | NaN |
| 1 | 200002.jpg | NaN |
| 2 | 200003.jpg | NaN |
| 3 | 200004.jpg | NaN |
| 4 | 200005.jpg | NaN |
Add a column with the full path to the images:
[10]:
%%time
train_data['path'] = INPUT_DIR + 'train_images/' + train_data['label'] + '/' + train_data['image_id']
train_data.head()
CPU times: user 4.89 ms, sys: 485 µs, total: 5.37 ms
Wall time: 5.14 ms
[10]:
| image_id | label | variety | age | path | |
|---|---|---|---|---|---|
| 0 | 100330.jpg | bacterial_leaf_blight | ADT45 | 45 | ./paddy-disease/train_images/bacterial_leaf_bl... |
| 1 | 100365.jpg | bacterial_leaf_blight | ADT45 | 45 | ./paddy-disease/train_images/bacterial_leaf_bl... |
| 2 | 100382.jpg | bacterial_leaf_blight | ADT45 | 45 | ./paddy-disease/train_images/bacterial_leaf_bl... |
| 3 | 100632.jpg | bacterial_leaf_blight | ADT45 | 45 | ./paddy-disease/train_images/bacterial_leaf_bl... |
| 4 | 101918.jpg | bacterial_leaf_blight | ADT45 | 45 | ./paddy-disease/train_images/bacterial_leaf_bl... |
[11]:
submission['path'] = INPUT_DIR + 'test_images/' + submission['image_id']
submission.head()
[11]:
| image_id | label | path | |
|---|---|---|---|
| 0 | 200001.jpg | NaN | ./paddy-disease/test_images/200001.jpg |
| 1 | 200002.jpg | NaN | ./paddy-disease/test_images/200002.jpg |
| 2 | 200003.jpg | NaN | ./paddy-disease/test_images/200003.jpg |
| 3 | 200004.jpg | NaN | ./paddy-disease/test_images/200004.jpg |
| 4 | 200005.jpg | NaN | ./paddy-disease/test_images/200005.jpg |
Let’s expand the training data with augmentations: random rotations and flips:
[ ]:
os.mkdir('./paddy-disease/modified_train')
[12]:
from PIL import Image
from tqdm.notebook import tqdm
new_imgs = []
for i, p in tqdm(enumerate(train_data['path'].values)):
if i % 1000 == 0:
print(i)
img = Image.open(p)
for it in range(10):
new_img = img.rotate(np.random.rand() * 60 - 30, resample=3)
if np.random.rand() > 0.5:
new_img = new_img.transpose(Image.FLIP_LEFT_RIGHT)
new_img_name = './paddy-disease/modified_train/' + p.split('/')[-1][:-4] + '_' + str(it) + '.jpg'
new_img.save(new_img_name)
new_imgs.append([new_img_name, p.split('/')[-2], p.split('/')[-1]])
0
1000
2000
3000
4000
5000
6000
7000
8000
9000
10000
[13]:
train_data = pd.concat([train_data, pd.DataFrame(new_imgs, columns = ['path', 'label', 'image_id'])]).reset_index(drop = True)
train_data
[13]:
| image_id | label | variety | age | path | |
|---|---|---|---|---|---|
| 0 | 100330.jpg | bacterial_leaf_blight | ADT45 | 45.0 | ./paddy-disease/train_images/bacterial_leaf_bl... |
| 1 | 100365.jpg | bacterial_leaf_blight | ADT45 | 45.0 | ./paddy-disease/train_images/bacterial_leaf_bl... |
| 2 | 100382.jpg | bacterial_leaf_blight | ADT45 | 45.0 | ./paddy-disease/train_images/bacterial_leaf_bl... |
| 3 | 100632.jpg | bacterial_leaf_blight | ADT45 | 45.0 | ./paddy-disease/train_images/bacterial_leaf_bl... |
| 4 | 101918.jpg | bacterial_leaf_blight | ADT45 | 45.0 | ./paddy-disease/train_images/bacterial_leaf_bl... |
| ... | ... | ... | ... | ... | ... |
| 114472 | 110381.jpg | tungro | NaN | NaN | ./paddy-disease/modified_train/110381_5.jpg |
| 114473 | 110381.jpg | tungro | NaN | NaN | ./paddy-disease/modified_train/110381_6.jpg |
| 114474 | 110381.jpg | tungro | NaN | NaN | ./paddy-disease/modified_train/110381_7.jpg |
| 114475 | 110381.jpg | tungro | NaN | NaN | ./paddy-disease/modified_train/110381_8.jpg |
| 114476 | 110381.jpg | tungro | NaN | NaN | ./paddy-disease/modified_train/110381_9.jpg |
114477 rows × 5 columns
Let’s do the same for the test dataset:
[22]:
os.mkdir('./paddy-disease/modified_test')
[14]:
new_imgs = []
for i, p in tqdm(enumerate(submission['path'].values)):
if i % 1000 == 0:
print(i)
img = Image.open(p)
for it in range(5):
new_img = img.rotate(np.random.rand() * 60 - 30, resample=3)
if np.random.rand() > 0.5:
new_img = new_img.transpose(Image.FLIP_LEFT_RIGHT)
new_img_name = './paddy-disease/modified_test/' + p.split('/')[-1][:-4] + '_' + str(it) + '.jpg'
new_img.save(new_img_name)
new_imgs.append([new_img_name, p.split('/')[-1]])
0
1000
2000
3000
[15]:
submission = pd.concat([submission, pd.DataFrame(new_imgs, columns = ['path', 'image_id'])]).reset_index(drop = True)
submission
[15]:
| image_id | label | path | |
|---|---|---|---|
| 0 | 200001.jpg | NaN | ./paddy-disease/test_images/200001.jpg |
| 1 | 200002.jpg | NaN | ./paddy-disease/test_images/200002.jpg |
| 2 | 200003.jpg | NaN | ./paddy-disease/test_images/200003.jpg |
| 3 | 200004.jpg | NaN | ./paddy-disease/test_images/200004.jpg |
| 4 | 200005.jpg | NaN | ./paddy-disease/test_images/200005.jpg |
| ... | ... | ... | ... |
| 20809 | 203469.jpg | NaN | ./paddy-disease/modified_test/203469_0.jpg |
| 20810 | 203469.jpg | NaN | ./paddy-disease/modified_test/203469_1.jpg |
| 20811 | 203469.jpg | NaN | ./paddy-disease/modified_test/203469_2.jpg |
| 20812 | 203469.jpg | NaN | ./paddy-disease/modified_test/203469_3.jpg |
| 20813 | 203469.jpg | NaN | ./paddy-disease/modified_test/203469_4.jpg |
20814 rows × 3 columns
Task definition
Task type
On the cell below we create Task object - the class to setup what task LightAutoML model should solve with specific loss and metric if necessary (more info can be found here in our documentation). In general, it can be any type of tasks available in LightAutoML (binary and multi-class classification, one-dimensional and multi-dimensional regression,
multi-label classification), but in this case we have a multi-class classification task:
[16]:
task = Task('multiclass')
Default metric and loss in multi-class classification is cross-entropy.
Feature roles setup
Next we need to setup columns roles. It is necessary to specify the role of the target variable ('target'), as well as the role of the path to the images ('path') in the case of using TabularCVAutoML. We will also group the images (the original ones and their augmentations) and apply group k-fold cross-validation, specifying the column with ids as the 'group' role:
[17]:
roles = {
'target': 'label',
'path': ['path'],
'drop': ['variety', 'age'],
'group': 'image_id'
}
Then we initialize TabularCVAutoML. It is possible to specify many parameters (reader parameters, time and memory limits etc), including the EfficientNet parameters for getting embeddings: version (B0 by default), device, batch size (128 by default), path for weights, AdvProp weights using (for better use of the shape in images, True by default) etc. Note that the Utilized version of TabularCVAutoML for more flexible use of time
resources is not yet available.
[18]:
automl = TabularCVAutoML(task = task,
timeout=5 * 3600,
cpu_limit = 2,
reader_params = {'cv': 5, 'random_state': 42})
AutoML training
To run autoML training use fit_predict method: - train_data - Dataset to train. - roles - Roles dict. - verbose - Controls the verbosity: the higher, the more messages. <1 : messages are not displayed; >=1 : the computation process for layers is displayed; >=2 : the information about folds processing is also displayed; >=3 : the hyperparameters optimization process is also displayed; >=4 : the training process for every algorithm is displayed;
Note: out-of-fold prediction is calculated during training and returned from the fit_predict method
[19]:
%%time
oof_pred = automl.fit_predict(train_data, roles = roles, verbose = 3)
[14:04:32] Stdout logging level is INFO3.
[14:04:32] Task: multiclass
[14:04:32] Start automl preset with listed constraints:
[14:04:32] - time: 18000.00 seconds
[14:04:32] - CPU: 2 cores
[14:04:32] - memory: 16 GB
[14:04:32] Train data shape: (114477, 5)
[14:04:32] Layer 1 train process start. Time left 17999.83 secs
100%|██████████| 895/895 [07:29<00:00, 1.99it/s]
[14:12:09] Feature path transformed
[14:12:16] Start fitting Lvl_0_Pipe_0_Mod_0_LinearL2 ...
[14:12:17] ===== Start working with fold 0 for Lvl_0_Pipe_0_Mod_0_LinearL2 =====
[14:12:26] Linear model: C = 1e-05 score = -0.9995305866945853
[14:12:32] Linear model: C = 5e-05 score = -0.6879959560713191
[14:12:38] Linear model: C = 0.0001 score = -0.5802952177399445
[14:12:45] Linear model: C = 0.0005 score = -0.3907926611544111
[14:12:51] Linear model: C = 0.001 score = -0.33425017155675657
[14:13:00] Linear model: C = 0.005 score = -0.2559518217619532
[14:13:07] Linear model: C = 0.01 score = -0.24141776919439237
[14:13:15] Linear model: C = 0.05 score = -0.2431661172897411
[14:13:23] Linear model: C = 0.1 score = -0.25925367786528475
[14:13:24] ===== Start working with fold 1 for Lvl_0_Pipe_0_Mod_0_LinearL2 =====
[14:13:32] Linear model: C = 1e-05 score = -0.9872444001968863
[14:13:39] Linear model: C = 5e-05 score = -0.6682540100549987
[14:13:45] Linear model: C = 0.0001 score = -0.5574685730009872
[14:13:51] Linear model: C = 0.0005 score = -0.3653461360638747
[14:13:58] Linear model: C = 0.001 score = -0.31059360297670363
[14:14:05] Linear model: C = 0.005 score = -0.2370436682635623
[14:14:14] Linear model: C = 0.01 score = -0.22495884629469698
[14:14:21] Linear model: C = 0.05 score = -0.23420873784566962
[14:14:29] Linear model: C = 0.1 score = -0.25263966927426823
[14:14:29] ===== Start working with fold 2 for Lvl_0_Pipe_0_Mod_0_LinearL2 =====
[14:14:37] Linear model: C = 1e-05 score = -0.9554531133528031
[14:14:43] Linear model: C = 5e-05 score = -0.640784196156178
[14:14:49] Linear model: C = 0.0001 score = -0.5345024606190905
[14:14:57] Linear model: C = 0.0005 score = -0.3546726337461952
[14:15:04] Linear model: C = 0.001 score = -0.30344210801693483
[14:15:12] Linear model: C = 0.005 score = -0.2331574262775805
[14:15:19] Linear model: C = 0.01 score = -0.22071779776854528
[14:15:28] Linear model: C = 0.05 score = -0.22603075278344578
[14:15:36] Linear model: C = 0.1 score = -0.24138537694410292
[14:15:36] ===== Start working with fold 3 for Lvl_0_Pipe_0_Mod_0_LinearL2 =====
[14:15:44] Linear model: C = 1e-05 score = -0.973115505822288
[14:15:51] Linear model: C = 5e-05 score = -0.6613476137718094
[14:15:56] Linear model: C = 0.0001 score = -0.5539538946164072
[14:16:04] Linear model: C = 0.0005 score = -0.3666276035478478
[14:16:10] Linear model: C = 0.001 score = -0.31130200709742806
[14:16:18] Linear model: C = 0.005 score = -0.2326339584928626
[14:16:25] Linear model: C = 0.01 score = -0.21658099282365262
[14:16:33] Linear model: C = 0.05 score = -0.21364841773406087
[14:16:42] Linear model: C = 0.1 score = -0.2256018292053085
[14:16:51] Linear model: C = 0.5 score = -0.2763179966937595
[14:16:51] ===== Start working with fold 4 for Lvl_0_Pipe_0_Mod_0_LinearL2 =====
[14:16:58] Linear model: C = 1e-05 score = -0.9531496536787142
[14:17:05] Linear model: C = 5e-05 score = -0.6270339670737181
[14:17:10] Linear model: C = 0.0001 score = -0.517302736118502
[14:17:17] Linear model: C = 0.0005 score = -0.331531311465719
[14:17:23] Linear model: C = 0.001 score = -0.27798570249468424
[14:17:32] Linear model: C = 0.005 score = -0.20448637290477473
[14:17:39] Linear model: C = 0.01 score = -0.19081673660070902
[14:17:47] Linear model: C = 0.05 score = -0.1923892363102242
[14:17:56] Linear model: C = 0.1 score = -0.20661581389305533
[14:17:56] Fitting Lvl_0_Pipe_0_Mod_0_LinearL2 finished. score = -0.21831477243925082
[14:17:56] Lvl_0_Pipe_0_Mod_0_LinearL2 fitting and predicting completed
[14:17:56] Time left 17195.98 secs
[14:22:15] Start fitting Lvl_0_Pipe_1_Mod_0_CatBoost ...
[14:22:16] ===== Start working with fold 0 for Lvl_0_Pipe_1_Mod_0_CatBoost =====
[14:22:16] 0: learn: 2.2636799 test: 2.2649649 best: 2.2649649 (0) total: 6.85ms remaining: 27.4s
[14:22:35] bestTest = 0.2436411292
[14:22:35] bestIteration = 3999
[14:22:35] ===== Start working with fold 1 for Lvl_0_Pipe_1_Mod_0_CatBoost =====
[14:22:36] 0: learn: 2.2634692 test: 2.2632526 best: 2.2632526 (0) total: 6.16ms remaining: 24.6s
[14:22:55] bestTest = 0.2658199543
[14:22:55] bestIteration = 3999
[14:22:56] ===== Start working with fold 2 for Lvl_0_Pipe_1_Mod_0_CatBoost =====
[14:22:56] 0: learn: 2.2631654 test: 2.2656298 best: 2.2656298 (0) total: 6.08ms remaining: 24.3s
[14:23:16] bestTest = 0.2753673319
[14:23:16] bestIteration = 3999
[14:23:16] ===== Start working with fold 3 for Lvl_0_Pipe_1_Mod_0_CatBoost =====
[14:23:17] 0: learn: 2.2645696 test: 2.2657045 best: 2.2657045 (0) total: 6.76ms remaining: 27s
[14:23:37] bestTest = 0.2738943611
[14:23:37] bestIteration = 3996
[14:23:37] Shrink model to first 3997 iterations.
[14:23:37] ===== Start working with fold 4 for Lvl_0_Pipe_1_Mod_0_CatBoost =====
[14:23:38] 0: learn: 2.2642805 test: 2.2644245 best: 2.2644245 (0) total: 5.84ms remaining: 23.4s
[14:23:57] bestTest = 0.2538460334
[14:23:57] bestIteration = 3999
[14:23:58] Fitting Lvl_0_Pipe_1_Mod_0_CatBoost finished. score = -0.2625123265864018
[14:23:58] Lvl_0_Pipe_1_Mod_0_CatBoost fitting and predicting completed
[14:23:58] Time left 16834.07 secs
[14:23:58] Layer 1 training completed.
[14:23:58] Blending: optimization starts with equal weights and score -0.1879588701291192
/home/dvladimirvasilyev/anaconda3/envs/myenv/lib/python3.8/site-packages/sklearn/metrics/_classification.py:2916: UserWarning: The y_pred values do not sum to one. Starting from 1.5 thiswill result in an error.
warnings.warn(
[14:23:59] Blending: iteration 0: score = -0.18573794844833624, weights = [0.63928086 0.36071914]
[14:23:59] Blending: iteration 1: score = -0.18573794844833624, weights = [0.63928086 0.36071914]
[14:23:59] Blending: no score update. Terminated
[14:23:59] Automl preset training completed in 1167.35 seconds
[14:23:59] Model description:
Final prediction for new objects (level 0) =
0.63928 * (5 averaged models Lvl_0_Pipe_0_Mod_0_LinearL2) +
0.36072 * (5 averaged models Lvl_0_Pipe_1_Mod_0_CatBoost)
CPU times: user 18min 40s, sys: 3min 1s, total: 21min 42s
Wall time: 19min 27s
Сonsider out-of-fold predictions on train data. In case of classification, LightAutoML returns class probabilities as an output.
[21]:
preds = train_data[['image_id', 'label']]
preds
[21]:
| image_id | label | |
|---|---|---|
| 0 | 100330.jpg | bacterial_leaf_blight |
| 1 | 100365.jpg | bacterial_leaf_blight |
| 2 | 100382.jpg | bacterial_leaf_blight |
| 3 | 100632.jpg | bacterial_leaf_blight |
| 4 | 101918.jpg | bacterial_leaf_blight |
| ... | ... | ... |
| 114472 | 110381.jpg | tungro |
| 114473 | 110381.jpg | tungro |
| 114474 | 110381.jpg | tungro |
| 114475 | 110381.jpg | tungro |
| 114476 | 110381.jpg | tungro |
114477 rows × 2 columns
[22]:
for i in range(10):
preds['pred_' + str(i)] = oof_pred.data[:,i]
preds
/tmp/ipykernel_12895/1432655611.py:2: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
preds['pred_' + str(i)] = oof_pred.data[:,i]
/tmp/ipykernel_12895/1432655611.py:2: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
preds['pred_' + str(i)] = oof_pred.data[:,i]
[22]:
| image_id | label | pred_0 | pred_1 | pred_2 | pred_3 | pred_4 | pred_5 | pred_6 | pred_7 | pred_8 | pred_9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 100330.jpg | bacterial_leaf_blight | 0.023245 | 0.315283 | 0.470886 | 0.002528 | 0.021895 | 0.007454 | 0.001554 | 0.157142 | 8.914904e-06 | 4.559626e-06 |
| 1 | 100365.jpg | bacterial_leaf_blight | 0.003717 | 0.011035 | 0.028317 | 0.000110 | 0.003178 | 0.000015 | 0.000131 | 0.953496 | 1.555987e-07 | 5.692390e-07 |
| 2 | 100382.jpg | bacterial_leaf_blight | 0.025734 | 0.095088 | 0.208473 | 0.000879 | 0.007030 | 0.003382 | 0.000142 | 0.659271 | 3.872871e-07 | 2.898941e-07 |
| 3 | 100632.jpg | bacterial_leaf_blight | 0.002876 | 0.542942 | 0.027466 | 0.000317 | 0.036005 | 0.000398 | 0.000082 | 0.389901 | 3.837710e-06 | 9.339438e-06 |
| 4 | 101918.jpg | bacterial_leaf_blight | 0.009988 | 0.033572 | 0.017635 | 0.000032 | 0.008310 | 0.000136 | 0.000041 | 0.930286 | 1.554736e-07 | 1.530466e-07 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 114472 | 110381.jpg | tungro | 0.001716 | 0.109143 | 0.020722 | 0.001495 | 0.845324 | 0.000177 | 0.021384 | 0.000027 | 6.304998e-06 | 6.075803e-06 |
| 114473 | 110381.jpg | tungro | 0.022644 | 0.137650 | 0.026389 | 0.004165 | 0.788036 | 0.001093 | 0.019688 | 0.000259 | 3.142513e-05 | 4.477663e-05 |
| 114474 | 110381.jpg | tungro | 0.016897 | 0.072329 | 0.010469 | 0.005554 | 0.789777 | 0.001240 | 0.103631 | 0.000060 | 1.301366e-05 | 2.972130e-05 |
| 114475 | 110381.jpg | tungro | 0.008637 | 0.114299 | 0.082281 | 0.003465 | 0.560001 | 0.000741 | 0.230260 | 0.000112 | 1.909918e-04 | 1.351225e-05 |
| 114476 | 110381.jpg | tungro | 0.004179 | 0.099988 | 0.008320 | 0.004660 | 0.822037 | 0.000663 | 0.059627 | 0.000318 | 1.922170e-04 | 1.441010e-05 |
114477 rows × 12 columns
We will average forecasts for images by their augmentations:
[23]:
preds = preds.groupby(['image_id', 'label']).mean().reset_index()
preds
[23]:
| image_id | label | pred_0 | pred_1 | pred_2 | pred_3 | pred_4 | pred_5 | pred_6 | pred_7 | pred_8 | pred_9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 100001.jpg | brown_spot | 0.001334 | 0.000791 | 0.002372 | 5.432664e-03 | 0.005328 | 0.978495 | 0.002519 | 0.003511 | 7.897679e-05 | 1.378119e-04 |
| 1 | 100002.jpg | normal | 0.978428 | 0.011744 | 0.001621 | 3.187062e-03 | 0.002579 | 0.000282 | 0.000156 | 0.001969 | 3.391063e-05 | 1.971700e-07 |
| 2 | 100003.jpg | hispa | 0.004639 | 0.002192 | 0.992883 | 1.573081e-07 | 0.000026 | 0.000037 | 0.000005 | 0.000218 | 1.920397e-07 | 1.528186e-07 |
| 3 | 100004.jpg | blast | 0.000259 | 0.982406 | 0.004401 | 7.787708e-03 | 0.002372 | 0.002163 | 0.000173 | 0.000115 | 3.223106e-04 | 4.848040e-07 |
| 4 | 100005.jpg | hispa | 0.010951 | 0.047475 | 0.829855 | 1.200308e-05 | 0.091933 | 0.000418 | 0.018967 | 0.000370 | 1.118553e-05 | 8.759866e-06 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 10402 | 110403.jpg | tungro | 0.001664 | 0.002167 | 0.007366 | 4.507852e-03 | 0.981122 | 0.000052 | 0.001666 | 0.001455 | 1.527430e-07 | 3.928369e-07 |
| 10403 | 110404.jpg | normal | 0.932484 | 0.002359 | 0.049850 | 1.244102e-05 | 0.011696 | 0.000593 | 0.002646 | 0.000304 | 4.828784e-05 | 7.773816e-06 |
| 10404 | 110405.jpg | dead_heart | 0.000192 | 0.000044 | 0.000152 | 9.994839e-01 | 0.000001 | 0.000025 | 0.000058 | 0.000003 | 1.957294e-06 | 3.789358e-05 |
| 10405 | 110406.jpg | blast | 0.000226 | 0.977683 | 0.000268 | 9.254745e-03 | 0.004962 | 0.000595 | 0.004523 | 0.001717 | 5.624577e-04 | 2.080105e-04 |
| 10406 | 110407.jpg | brown_spot | 0.000009 | 0.000188 | 0.000539 | 4.357956e-04 | 0.000232 | 0.997215 | 0.000039 | 0.000010 | 1.319862e-03 | 1.372061e-05 |
10407 rows × 12 columns
Assign classes by maximum class probability:
[24]:
OOFs = np.argmax(preds[['pred_' + str(i) for i in range(10)]].values, axis = 1)
OOFs
[24]:
array([5, 0, 2, ..., 3, 1, 5])
Let’s see classification accuracy on train:
[25]:
accuracy = (OOFs == preds['label'].map(automl.reader.class_mapping)).mean()
print(f'Out-of-fold accuracy: {accuracy}')
Out-of-fold accuracy: 0.9686749303353512
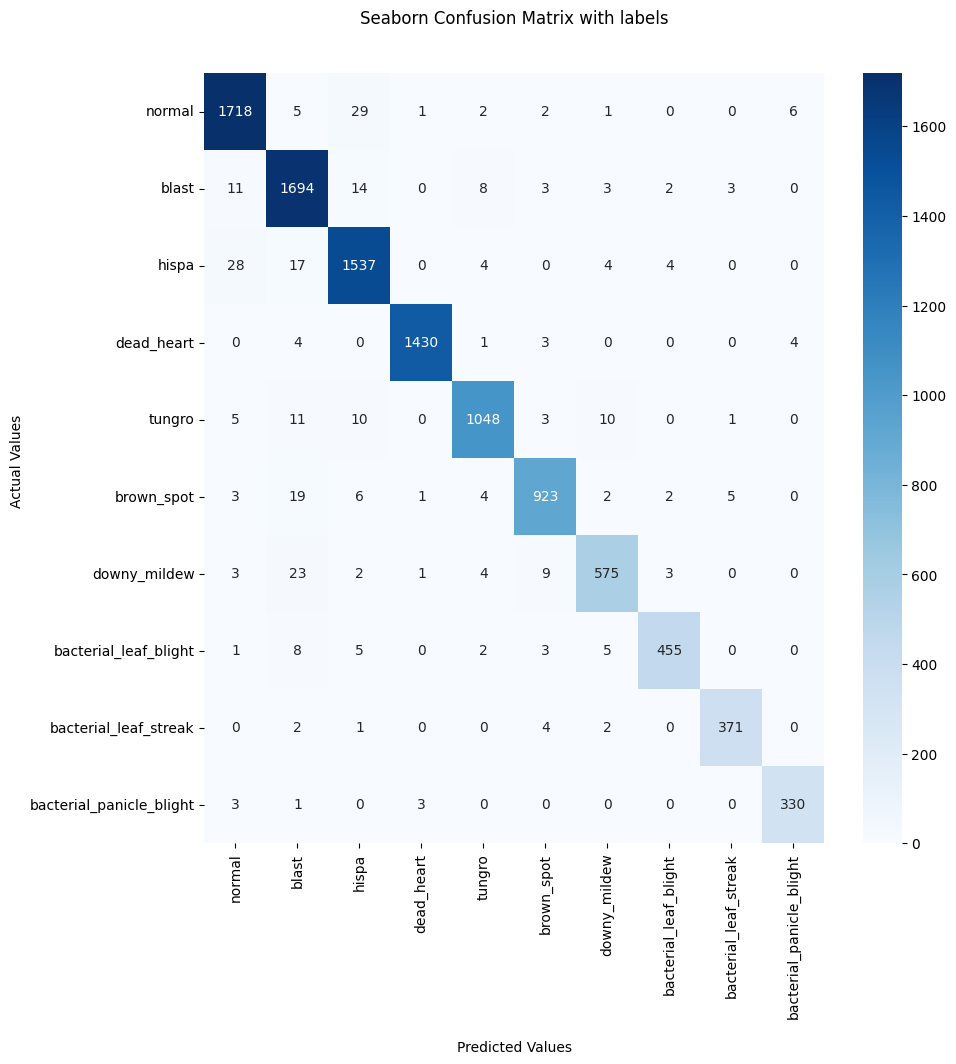
Also to estimate the quality of classification, we can use the confusion matrix:
[26]:
cf_matrix = confusion_matrix(preds['label'].map(automl.reader.class_mapping),
OOFs)
plt.figure(figsize = (10, 10))
ax = sns.heatmap(cf_matrix, annot=True, cmap='Blues', fmt = 'd')
ax.set_title('Seaborn Confusion Matrix with labels\n\n');
ax.set_xlabel('\nPredicted Values')
ax.set_ylabel('Actual Values ');
inverse_class_mapping = {y: x for x,y in automl.reader.class_mapping.items()}
labels = [inverse_class_mapping[i] for i in range(len(inverse_class_mapping))]
ax.xaxis.set_ticklabels(labels, rotation = 90)
ax.yaxis.set_ticklabels(labels, rotation = 0)
plt.show()
Predict for test dataset
Now we are also ready to predict for our test competition dataset and submission file creation:
[27]:
%%time
te_pred = automl.predict(submission)
print(f'Prediction for te_data:\n{te_pred}\nShape = {te_pred.shape}')
100%|██████████| 163/163 [01:28<00:00, 1.84it/s]
[14:28:22] Feature path transformed
Prediction for te_data:
array([[1.57098308e-01, 2.81519257e-03, 5.96348643e-01, ...,
1.08084995e-02, 1.95845146e-07, 1.42198633e-05],
[9.83384371e-01, 6.52049668e-04, 1.45791359e-02, ...,
1.12365209e-03, 9.75986836e-07, 1.95965598e-07],
[1.68020770e-01, 3.79674375e-01, 1.86414778e-01, ...,
1.67078048e-03, 1.21877249e-03, 3.75247910e-03],
...,
[1.05072348e-03, 1.24680300e-05, 5.70231769e-03, ...,
4.37476301e-05, 1.52421890e-07, 1.81421214e-07],
[6.52685121e-04, 4.47798493e-06, 5.04824053e-03, ...,
2.13344283e-05, 1.52417726e-07, 1.62638599e-07],
[1.57185504e-03, 1.01540554e-05, 2.53849756e-02, ...,
1.17763964e-04, 1.52426963e-07, 1.77946404e-07]], dtype=float32)
Shape = (20814, 10)
CPU times: user 55.8 s, sys: 21.6 s, total: 1min 17s
Wall time: 2min 19s
[28]:
sub = submission[['image_id']]
for i in range(10):
sub['pred_' + str(i)] = te_pred.data[:,i]
sub
/tmp/ipykernel_12895/1185757098.py:3: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
sub['pred_' + str(i)] = te_pred.data[:,i]
[28]:
| image_id | pred_0 | pred_1 | pred_2 | pred_3 | pred_4 | pred_5 | pred_6 | pred_7 | pred_8 | pred_9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 200001.jpg | 0.157098 | 0.002815 | 0.596349 | 0.020590 | 1.148577e-01 | 0.095614 | 0.001854 | 0.010808 | 1.958451e-07 | 1.421986e-05 |
| 1 | 200002.jpg | 0.983384 | 0.000652 | 0.014579 | 0.000139 | 6.825896e-05 | 0.000044 | 0.000008 | 0.001124 | 9.759868e-07 | 1.959656e-07 |
| 2 | 200003.jpg | 0.168021 | 0.379674 | 0.186415 | 0.000225 | 1.850213e-03 | 0.036919 | 0.220253 | 0.001671 | 1.218772e-03 | 3.752479e-03 |
| 3 | 200004.jpg | 0.000013 | 0.990730 | 0.008530 | 0.000097 | 1.116415e-04 | 0.000215 | 0.000111 | 0.000037 | 1.548404e-04 | 1.946677e-07 |
| 4 | 200005.jpg | 0.000340 | 0.999536 | 0.000031 | 0.000002 | 6.857538e-07 | 0.000003 | 0.000007 | 0.000029 | 5.404088e-07 | 4.985940e-05 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 20809 | 203469.jpg | 0.003061 | 0.000017 | 0.041731 | 0.943745 | 1.648944e-04 | 0.010877 | 0.000146 | 0.000258 | 1.524480e-07 | 2.509265e-07 |
| 20810 | 203469.jpg | 0.000430 | 0.000003 | 0.002508 | 0.993409 | 2.613632e-05 | 0.003580 | 0.000007 | 0.000036 | 1.524176e-07 | 1.595918e-07 |
| 20811 | 203469.jpg | 0.001051 | 0.000012 | 0.005702 | 0.989972 | 5.734707e-05 | 0.003144 | 0.000018 | 0.000044 | 1.524219e-07 | 1.814212e-07 |
| 20812 | 203469.jpg | 0.000653 | 0.000004 | 0.005048 | 0.990724 | 3.223727e-05 | 0.003505 | 0.000012 | 0.000021 | 1.524177e-07 | 1.626386e-07 |
| 20813 | 203469.jpg | 0.001572 | 0.000010 | 0.025385 | 0.965282 | 1.030424e-04 | 0.007472 | 0.000058 | 0.000118 | 1.524270e-07 | 1.779464e-07 |
20814 rows × 11 columns
[29]:
sub = sub.groupby(['image_id']).mean().reset_index()
sub
[29]:
| image_id | pred_0 | pred_1 | pred_2 | pred_3 | pred_4 | pred_5 | pred_6 | pred_7 | pred_8 | pred_9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 200001.jpg | 0.127650 | 0.001409 | 0.599914 | 0.017568 | 0.136898 | 0.106915 | 0.001796 | 0.007829 | 8.801418e-06 | 1.216593e-05 |
| 1 | 200002.jpg | 0.937035 | 0.000638 | 0.060420 | 0.000098 | 0.000105 | 0.000096 | 0.000016 | 0.001586 | 6.087082e-06 | 2.249314e-07 |
| 2 | 200003.jpg | 0.120163 | 0.523312 | 0.106169 | 0.000473 | 0.000748 | 0.042688 | 0.201373 | 0.002807 | 1.389023e-03 | 8.788786e-04 |
| 3 | 200004.jpg | 0.000020 | 0.888623 | 0.006415 | 0.001150 | 0.000430 | 0.004390 | 0.000616 | 0.001799 | 9.654120e-02 | 1.466518e-05 |
| 4 | 200005.jpg | 0.000680 | 0.998898 | 0.000085 | 0.000009 | 0.000001 | 0.000002 | 0.000021 | 0.000172 | 1.743805e-06 | 1.304403e-04 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 3464 | 203465.jpg | 0.000224 | 0.002143 | 0.001514 | 0.990281 | 0.002657 | 0.000401 | 0.001074 | 0.000134 | 1.530934e-03 | 4.091801e-05 |
| 3465 | 203466.jpg | 0.250769 | 0.007148 | 0.741840 | 0.000002 | 0.000022 | 0.000013 | 0.000076 | 0.000129 | 2.629060e-07 | 2.120279e-07 |
| 3466 | 203467.jpg | 0.960745 | 0.004105 | 0.001135 | 0.000646 | 0.016724 | 0.008584 | 0.000062 | 0.007749 | 2.438365e-04 | 6.326832e-06 |
| 3467 | 203468.jpg | 0.003675 | 0.001097 | 0.038018 | 0.000038 | 0.000483 | 0.000310 | 0.000223 | 0.000208 | 9.551883e-01 | 7.596347e-04 |
| 3468 | 203469.jpg | 0.001372 | 0.000012 | 0.015432 | 0.977533 | 0.000086 | 0.005415 | 0.000046 | 0.000104 | 1.524300e-07 | 1.962799e-07 |
3469 rows × 11 columns
[30]:
TEs = pd.Series(np.argmax(sub[['pred_' + str(i) for i in range(10)]].values, axis = 1)).map(inverse_class_mapping)
TEs
[30]:
0 hispa
1 normal
2 blast
3 blast
4 blast
...
3464 dead_heart
3465 hispa
3466 normal
3467 bacterial_leaf_streak
3468 dead_heart
Length: 3469, dtype: object
[31]:
sub['label'] = TEs
sub[['image_id', 'label']].to_csv('LightAutoML_TabularCVAutoML_with_aug.csv', index = False)
sub[['image_id', 'label']]
[31]:
| image_id | label | |
|---|---|---|
| 0 | 200001.jpg | hispa |
| 1 | 200002.jpg | normal |
| 2 | 200003.jpg | blast |
| 3 | 200004.jpg | blast |
| 4 | 200005.jpg | blast |
| ... | ... | ... |
| 3464 | 203465.jpg | dead_heart |
| 3465 | 203466.jpg | hispa |
| 3466 | 203467.jpg | normal |
| 3467 | 203468.jpg | bacterial_leaf_streak |
| 3468 | 203469.jpg | dead_heart |
3469 rows × 2 columns
No we can choose another model from timm. So we will use resnet50.a1_in1k, by default it uses vit_base_patch16_224.augreg_in21k
[35]:
automl = TabularCVAutoML(task = task,
timeout=5 * 3600,
autocv_features={"embed_model": 'timm/tf_efficientnetv2_b0.in1k'},
cpu_limit = 2,
reader_params = {'cv': 5, 'random_state': 42})
[36]:
%%time
oof_pred = automl.fit_predict(train_data, roles = roles, verbose = 3)
[14:37:43] Stdout logging level is INFO3.
[14:37:43] Task: multiclass
[14:37:43] Start automl preset with listed constraints:
[14:37:43] - time: 18000.00 seconds
[14:37:43] - CPU: 2 cores
[14:37:43] - memory: 16 GB
[14:37:43] Train data shape: (114477, 5)
[14:37:43] Layer 1 train process start. Time left 17999.80 secs
100%|██████████| 895/895 [06:43<00:00, 2.22it/s]
[14:44:31] Feature path transformed
[14:44:41] Start fitting Lvl_0_Pipe_0_Mod_0_LinearL2 ...
[14:44:41] ===== Start working with fold 0 for Lvl_0_Pipe_0_Mod_0_LinearL2 =====
[14:44:53] Linear model: C = 1e-05 score = -1.2282992628176856
[14:45:04] Linear model: C = 5e-05 score = -0.9078946864858105
[14:45:14] Linear model: C = 0.0001 score = -0.7903223383077203
[14:45:25] Linear model: C = 0.0005 score = -0.5805263796419443
[14:45:37] Linear model: C = 0.001 score = -0.5191830537228186
[14:45:48] Linear model: C = 0.005 score = -0.44237800607788724
[14:46:01] Linear model: C = 0.01 score = -0.4332587963951451
[14:46:16] Linear model: C = 0.05 score = -0.4659824021930572
[14:46:28] Linear model: C = 0.1 score = -0.49696980356910764
[14:46:29] ===== Start working with fold 1 for Lvl_0_Pipe_0_Mod_0_LinearL2 =====
[14:46:40] Linear model: C = 1e-05 score = -1.1941203869888553
[14:46:50] Linear model: C = 5e-05 score = -0.870315687726058
[14:47:00] Linear model: C = 0.0001 score = -0.7542737074009194
[14:47:11] Linear model: C = 0.0005 score = -0.5565397834768919
[14:47:23] Linear model: C = 0.001 score = -0.5021799803891854
[14:47:37] Linear model: C = 0.005 score = -0.4375446715586552
[14:47:49] Linear model: C = 0.01 score = -0.4337117229695793
[14:48:03] Linear model: C = 0.05 score = -0.47678539878379567
[14:48:16] Linear model: C = 0.1 score = -0.5100193461879381
[14:48:16] ===== Start working with fold 2 for Lvl_0_Pipe_0_Mod_0_LinearL2 =====
[14:48:27] Linear model: C = 1e-05 score = -1.1828501053814764
[14:48:39] Linear model: C = 5e-05 score = -0.8603329618510173
[14:48:48] Linear model: C = 0.0001 score = -0.7451147263666518
[14:48:59] Linear model: C = 0.0005 score = -0.5469582228988039
[14:49:12] Linear model: C = 0.001 score = -0.49160247842297417
[14:49:24] Linear model: C = 0.005 score = -0.4257572256164155
[14:49:37] Linear model: C = 0.01 score = -0.4188241529929714
[14:49:50] Linear model: C = 0.05 score = -0.4522382557188784
[14:50:03] Linear model: C = 0.1 score = -0.48277984079191094
[14:50:04] ===== Start working with fold 3 for Lvl_0_Pipe_0_Mod_0_LinearL2 =====
[14:50:15] Linear model: C = 1e-05 score = -1.1958343845422246
[14:50:26] Linear model: C = 5e-05 score = -0.878725101433787
[14:50:35] Linear model: C = 0.0001 score = -0.7660166437189271
[14:50:45] Linear model: C = 0.0005 score = -0.5679153687919936
[14:50:59] Linear model: C = 0.001 score = -0.5110457138416219
[14:51:10] Linear model: C = 0.005 score = -0.44229320617124224
[14:51:23] Linear model: C = 0.01 score = -0.43663952743918066
[14:51:37] Linear model: C = 0.05 score = -0.47363171137894655
[14:51:51] Linear model: C = 0.1 score = -0.5032655687259646
[14:51:51] ===== Start working with fold 4 for Lvl_0_Pipe_0_Mod_0_LinearL2 =====
[14:52:02] Linear model: C = 1e-05 score = -1.1804715353776323
[14:52:13] Linear model: C = 5e-05 score = -0.8529105474280552
[14:52:21] Linear model: C = 0.0001 score = -0.7373622302487922
[14:52:32] Linear model: C = 0.0005 score = -0.537561225715503
[14:52:43] Linear model: C = 0.001 score = -0.48106564988541606
[14:52:57] Linear model: C = 0.005 score = -0.4138154861612588
[14:53:09] Linear model: C = 0.01 score = -0.40990101492044817
[14:53:23] Linear model: C = 0.05 score = -0.44904189928940963
[14:53:36] Linear model: C = 0.1 score = -0.4789966864522385
[14:53:36] Fitting Lvl_0_Pipe_0_Mod_0_LinearL2 finished. score = -0.4264683916927181
[14:53:36] Lvl_0_Pipe_0_Mod_0_LinearL2 fitting and predicting completed
[14:53:36] Time left 17046.53 secs
[14:58:02] Start fitting Lvl_0_Pipe_1_Mod_0_CatBoost ...
[14:58:02] ===== Start working with fold 0 for Lvl_0_Pipe_1_Mod_0_CatBoost =====
[14:58:02] 0: learn: 2.2636799 test: 2.2649651 best: 2.2649651 (0) total: 10.4ms remaining: 41.6s
[14:58:22] bestTest = 0.2436411292
[14:58:22] bestIteration = 3999
[14:58:23] ===== Start working with fold 1 for Lvl_0_Pipe_1_Mod_0_CatBoost =====
[14:58:23] 0: learn: 2.2634693 test: 2.2632523 best: 2.2632523 (0) total: 6.07ms remaining: 24.3s
[14:58:43] bestTest = 0.2658199756
[14:58:43] bestIteration = 3999
[14:58:43] ===== Start working with fold 2 for Lvl_0_Pipe_1_Mod_0_CatBoost =====
[14:58:44] 0: learn: 2.2631659 test: 2.2656305 best: 2.2656305 (0) total: 6.52ms remaining: 26.1s
[14:59:03] bestTest = 0.2753673959
[14:59:03] bestIteration = 3999
[14:59:04] ===== Start working with fold 3 for Lvl_0_Pipe_1_Mod_0_CatBoost =====
[14:59:04] 0: learn: 2.2645703 test: 2.2657044 best: 2.2657044 (0) total: 6.13ms remaining: 24.5s
[14:59:24] bestTest = 0.2738942971
[14:59:24] bestIteration = 3996
[14:59:24] Shrink model to first 3997 iterations.
[14:59:24] ===== Start working with fold 4 for Lvl_0_Pipe_1_Mod_0_CatBoost =====
[14:59:25] 0: learn: 2.2642798 test: 2.2644247 best: 2.2644247 (0) total: 5.95ms remaining: 23.8s
[14:59:44] bestTest = 0.2538460547
[14:59:44] bestIteration = 3999
[14:59:45] Fitting Lvl_0_Pipe_1_Mod_0_CatBoost finished. score = -0.2625123265864018
[14:59:45] Lvl_0_Pipe_1_Mod_0_CatBoost fitting and predicting completed
[14:59:45] Time left 16678.32 secs
[14:59:45] Layer 1 training completed.
[14:59:45] Blending: optimization starts with equal weights and score -0.2561708318332855
/home/dvladimirvasilyev/anaconda3/envs/myenv/lib/python3.8/site-packages/sklearn/metrics/_classification.py:2916: UserWarning: The y_pred values do not sum to one. Starting from 1.5 thiswill result in an error.
warnings.warn(
[14:59:45] Blending: iteration 0: score = -0.23692344794948073, weights = [0.19089036 0.8091096 ]
[14:59:46] Blending: iteration 1: score = -0.23692344794948073, weights = [0.19089036 0.8091096 ]
[14:59:46] Blending: no score update. Terminated
[14:59:46] Automl preset training completed in 1323.26 seconds
[14:59:46] Model description:
Final prediction for new objects (level 0) =
0.19089 * (5 averaged models Lvl_0_Pipe_0_Mod_0_LinearL2) +
0.80911 * (5 averaged models Lvl_0_Pipe_1_Mod_0_CatBoost)
CPU times: user 20min 56s, sys: 1min 25s, total: 22min 22s
Wall time: 22min 3s
[37]:
%%time
te_pred = automl.predict(submission)
print(f'Prediction for te_data:\n{te_pred}\nShape = {te_pred.shape}')
100%|██████████| 163/163 [01:16<00:00, 2.13it/s]
[15:01:03] Feature path transformed
Prediction for te_data:
array([[5.8534566e-02, 6.8576052e-03, 4.5334366e-01, ..., 1.5735241e-02,
4.2415738e-07, 2.8625556e-05],
[9.6386713e-01, 1.4697504e-03, 3.2047924e-02, ..., 2.0407902e-03,
6.7228694e-07, 1.4319470e-07],
[3.5120246e-01, 2.9431397e-01, 1.9644174e-01, ..., 2.2667376e-04,
6.4593733e-06, 5.9228983e-05],
...,
[2.3565248e-03, 2.7670001e-05, 1.2790265e-02, ..., 9.7831573e-05,
4.5524594e-08, 1.1057142e-07],
[1.4637065e-03, 9.7479615e-06, 1.1323140e-02, ..., 4.7557736e-05,
4.5515264e-08, 6.8441139e-08],
[3.5254466e-03, 2.2519611e-05, 5.6939691e-02, ..., 2.6386546e-04,
4.5536019e-08, 1.7701051e-07]], dtype=float32)
Shape = (20814, 10)
CPU times: user 13 s, sys: 3.3 s, total: 16.3 s
Wall time: 2min 7s
Our submission has 0.95770 accuracy on public and 0.95276 accuracy on private leaderboard (Alexander Ryzhkov account).